Abstract
Introduction: Current data suggests Black/African Americans (AA) have an increased risk of developing multiple myeloma (MM), especially in comparison to White/European Americans (EA). However, the underlying causes remain unclear. Genetic definition of the spectrum of acquired mutations and the signatures they contain could help us to understand the mechanism underlying this excess risk. Currently, only limited information is available on the mutational spectra within AAs and the majority of data that has been generated is based on the societal construct of self-identified race/ethnicity alone. To address this deficiency and the potential associated bias, it is critical to examine greater numbers of whole genome sequencing (WGS) data from AAs. Data from a pilot study indicate a prolonged and more intense exposure to the germinal center (GC) reaction in the early evolutionary phases of MM within AAs, suggesting that it is essential to examine sequencing data from early disease stage i.e. smoldering MM (SMM) to better understand genetic predisposition in AAs.
Methods: To address the mutational basis of MM and how it varies based on racial origin we carried out two large studies, Polyethnic-1000 and the Smoldering Myeloma Registration Trial (SMRT) study. Furthermore, we examined a cross-sectional series of WGS and whole exome sequencing (WES) data derived from cases with SMM and MM already sequenced to characterize the pattern of mutations and the signatures in both the coding and non-coding regions of the genome. Importantly, to accurately infer racial ancestry, we built an admixture workflow into our sequencing analysis pipeline, enabling us to address the variability in origin from within the MM cases ranging from regions of Europe to subsets of cases with African, North/South American, and Asian origin directly. We showed its use in previous work describing the heterogenous genetic admixture in MM cases that self-identified Hispanic and Latino.
Our pipeline uses a suite of well-supported bioinformatic tools for preprocessing sequencing data according to best practices, cataloguing germline variants across a large series of patients, generating ancestry admixture estimations per patient, determining telomere length and composition, and identifying the spectrum of somatic events including single nucleotide variants, small insertions/deletions, copy number variants, and structural variants using consensus calling. All source code and reference data is publicly available via a GitHub repository (https://github.com/pblaney/mgp1000).
Results: We generated new WGS data from 65 MM cases (EA n=15, AA n=50), and 44 SMM cases (EA n=36, AA n=18) as part of the Polyethnic 1000 and SMRT study. We also systematically reanalyzed 473 cases with deep (>30x) WGS data and all cases in the CoMMpass study low-depth (5-12x) WGS (n=943).
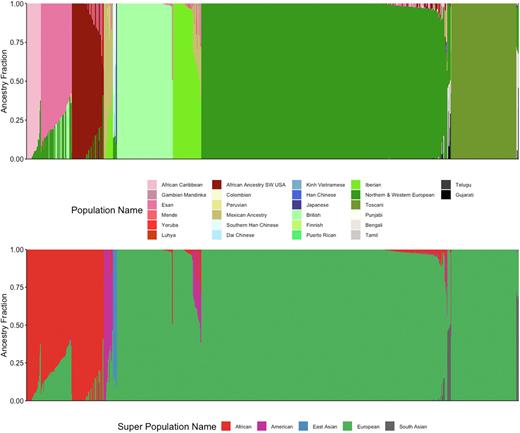
Estimating ancestral composition directly from the genetic data of the CoMMpass study, we identified discrepancies between the self-reported race and the genetically determined admixture composition. Approximately 75% of individuals identified as 'White' but 7% of these cases had a genetic composition that included greater than 10% contribution from African, North/South American, or Asian ancestry. More interesting, 18% identified as 'Black/African American' but 50% of these cases had greater than 10% contribution from European, North/South American, or Asian ancestry. These results demonstrate the underlying genetic heterogeneity in a population that is currently clinically classified as the homogenous and thus lead to skewed evaluations of race and outcome in MM.
We will compare the mutational spectra within these groups with different origins to address the risk of developing MM as well as addressing the time of origin of MM within the different populations.
Conclusion: We show that there is significant variability of admixture from Europe, North/South America, and Africa within patients who self-identify as White, Black/African American, or Hispanic/Latino in clinical datasets from the U.S. This reinforces the need for complementary genetic admixture with racial self-identification when investigating the links between racial ancestry and outcomes in MM.
Figure: Admixture composition of CoMMpass study shows significant contribution of African, North/South American, and East/South Asian origins in predominant European ancestral background.
Disclosures
Braunstein:Abbvie: Membership on an entity's Board of Directors or advisory committees; ADC Therapeutics: Membership on an entity's Board of Directors or advisory committees; Amgen: Membership on an entity's Board of Directors or advisory committees; BMS: Membership on an entity's Board of Directors or advisory committees; Epizyme: Membership on an entity's Board of Directors or advisory committees; GSK: Membership on an entity's Board of Directors or advisory committees; Janssen: Membership on an entity's Board of Directors or advisory committees; Morphosys: Membership on an entity's Board of Directors or advisory committees; Pfizer: Membership on an entity's Board of Directors or advisory committees; TG Therapeutics: Membership on an entity's Board of Directors or advisory committees. Diamond:Sanofi: Honoraria; Janssen: Honoraria; Medscape: Honoraria. Kazandjian:SINTOMA: Honoraria; Sanofi: Membership on an entity's Board of Directors or advisory committees; Arcellx: Membership on an entity's Board of Directors or advisory committees; MMRF: Honoraria; Curio Science: Honoraria; Aptitude Health: Honoraria; Plexus Communications: Honoraria; BMS: Membership on an entity's Board of Directors or advisory committees; CURE: Honoraria. Shah:MashUpMD: Honoraria; Sanofi: Consultancy; ACCC: Honoraria; Janssen: Consultancy, Research Funding; Bristol Myers Squibb: Consultancy, Research Funding; MJH Lifesciences: Consultancy, Honoraria. Lesokhin:Amgen: Honoraria; BMS: Honoraria; Sanofi: Research Funding; Trillium Therapeutics: Consultancy, Research Funding; Serametrix, inc: Patents & Royalties; Janssen, Pfizer, Iteos, Sanofi, Genmab: Honoraria; Janssen, Pfizer, BMS, Genentech/Roche: Research Funding; Pfizer, Genmab, Sanofi, Iteos, BMS, Janssen: Consultancy; Memorial Sloan Kettering Cancer Center: Current Employment. Walker:Genentech: Research Funding; Bristol Myers Squibb: Research Funding. Usmani:Amgen, BMS, Janssen, Sanofi: Speakers Bureau; Abbvie, Amgen, BMS, Celgene, EdoPharma, Genentech, Gilead, GSK, Janssen,Oncopeptides, Sanofi, Seattle Genetics, SecuraBio, SkylineDX, Takeda, TeneoBio: Consultancy; Amgen, Array Biopharma, BMS, Celgene, GSK, Janssen, Merck, Pharmacyclics, Sanofi, Seattle Genetics, SkylineDX, Takeda: Research Funding. Landgren:Janssen: Honoraria, Other: Independent Data Monitoring Committee (IDMC) member for clinical trials, Research Funding; Merck & Co., Inc.: Other: Independent Data Monitoring Committee (IDMC) member for clinical trials; Tow Foundation: Research Funding; Rising Tide Foundation: Research Funding; Leukemia & Lymphoma Society: Research Funding; MMRF: Honoraria; Aptitude Health: Honoraria; NCI/NIH: Research Funding; Riney Foundation: Research Funding; Pfizer Inc: Consultancy; Theradex: Other: Independent Data Monitoring Committee (IDMC) member for clinical trials; Amgen: Honoraria, Research Funding. Davies:Roche: Consultancy, Honoraria; Takeda: Consultancy, Honoraria; Janssen: Consultancy, Honoraria; Amgen: Consultancy, Honoraria; Abbvie: Consultancy, Honoraria; Celgene/BMS: Consultancy, Honoraria.
Author notes
Asterisk with author names denotes non-ASH members.